Script conventions
MLReef is developed to be non-intrusive as possible. There are no dedicated libraries necessary to run your code and all your scripts will perfectly work outside of MLReef.
To enable this openess and also to no limit your usage, we had to implement some conventions in regards to your scripts. These are the only conventions required:
| Convention | Documentation for |
|---|---|
| Argparse params & values | All scripts require argparse. |
| Data Input & Output | Each script needs an input and output parameter. |
| Entry point | To publish, you need one entry point. |
| Experiment metrics | Your models need to output a experiment.json file for metrics. |
Argparse params & values
You can only publish scripts into AI Modules if your entry point script or params.py file uses Argparse to annotate your parameters.
What does argparse do?
The argparse module makes it easy to write user-friendly command-line interfaces. The program defines what arguments it requires, and argparse will figure out how to parse those out of sys.argv. The argparse module also automatically generates help and usage messages. The module will also issue errors when users give the program invalid arguments.
It should look something like this:
def process_arguments(args):
parser = argparse.ArgumentParser(description='ResNet50')
parser.add_argument('--input-path', action='store', required=true, help='path to directory of images')
parser.add_argument('--output-path', action='store', required=true,default='.', help='path to output metrics')
parser.add_argument('--height', action='store', default=224, required=true, help='height of images (int)')
parser.add_argument('--width', action='store', default=224,required=true, help='width of images (int)')
arser.add_argument('--channels', action='store', default=3, help='channels of images: 1 = grayscale, 3 = RGB , 4=RGBA (int)')
params = vars(parser.parse_args(args))
return params
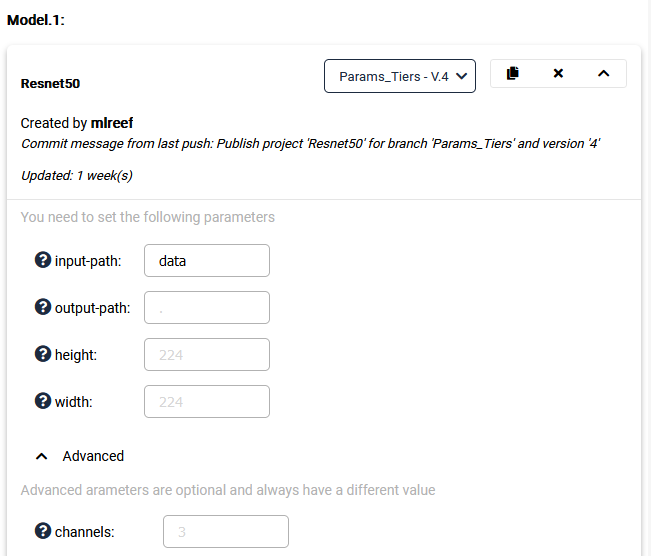
<> Note: Some arguments will have a decorator "required=true". You can use this to mark params that require user input during pipeline creation and only "advanced" parameters that have a default value.
This will then look like this:
Data Input & Output
Your script needs to contain at least a function to load (input) and write (output) data. We mostly use a param "input-path" and "output-path" to define the source of your data to use in the pipelines and also where to store any output created.
Remember to include at least both in your script:
def process_arguments(args):
parser = argparse.ArgumentParser(description='ResNet50')
parser.add_argument('--input-path', action='store', required=true, help='path to directory of images')
parser.add_argument('--output-path', action='store', required=true,default='.', help='path to output metrics')
Another tip: Use functions that create directories in your script. This will help you organize your output.
Entry point
The publishing process requires a single entry point. This can be a main.py file or, if used on a singular script, your script. During publishing, this entry point will be parsed to read out your argparse marked parameters.
Most importantly, this script will be executed during your pipeline run via command line. The command will look like this:
python run ResNet50 --input-path /folder --output-path /output --width 200 --height 200 --channels 3
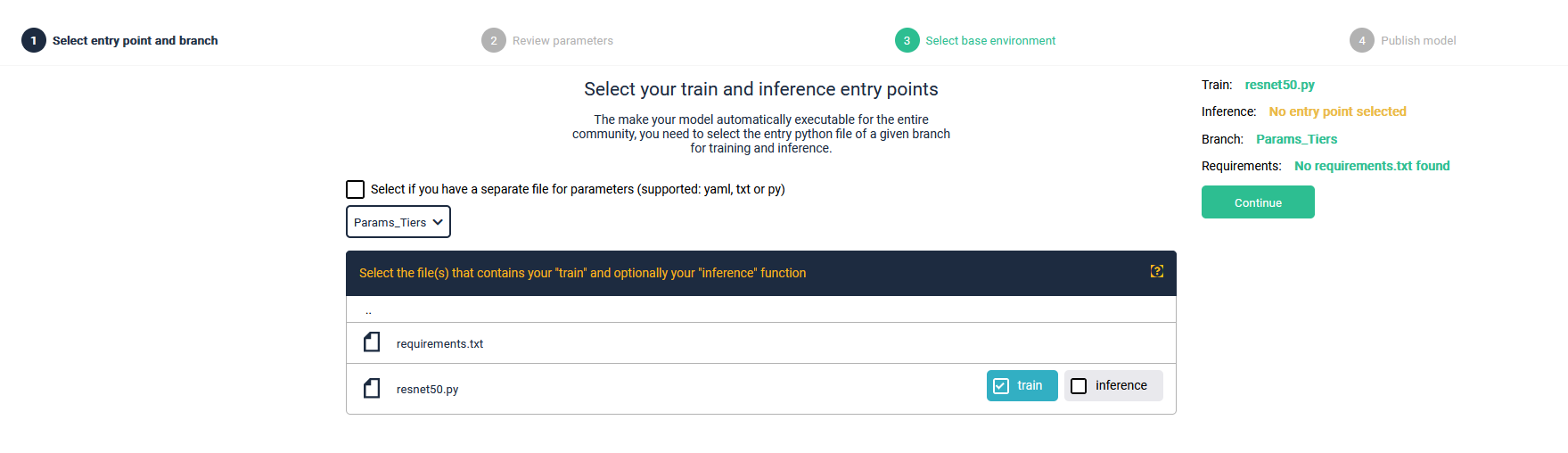
If your script contains any other parameters in other scripts other than your entry point, then these will not be included in your AI Module!
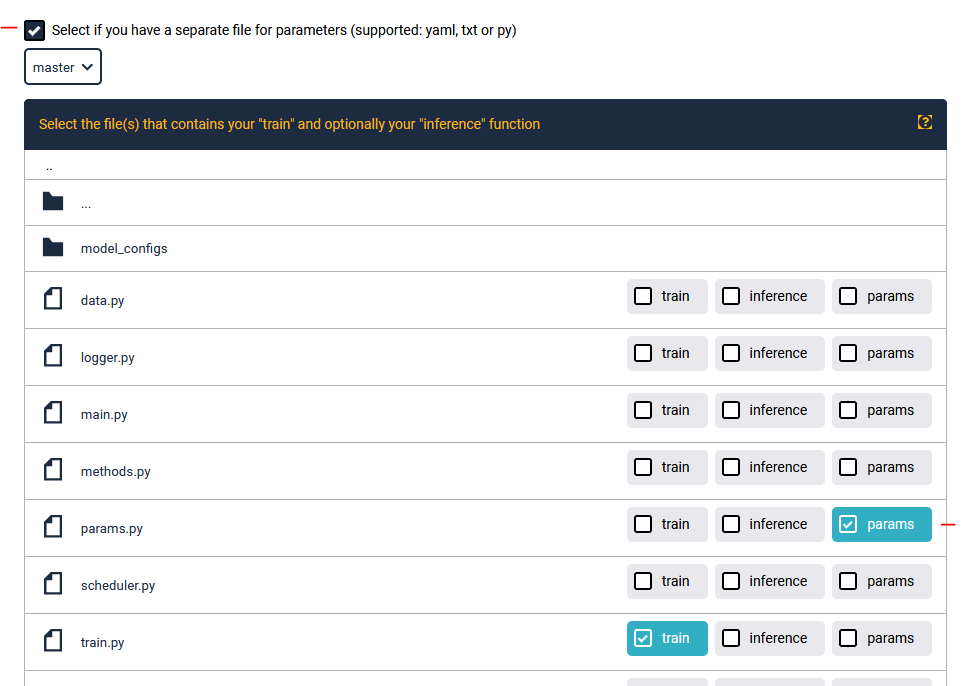
You can also specify a dedicated params file containing all addressable arguments. During publishing, simply select this option and mark your params file:
Experiment metrics
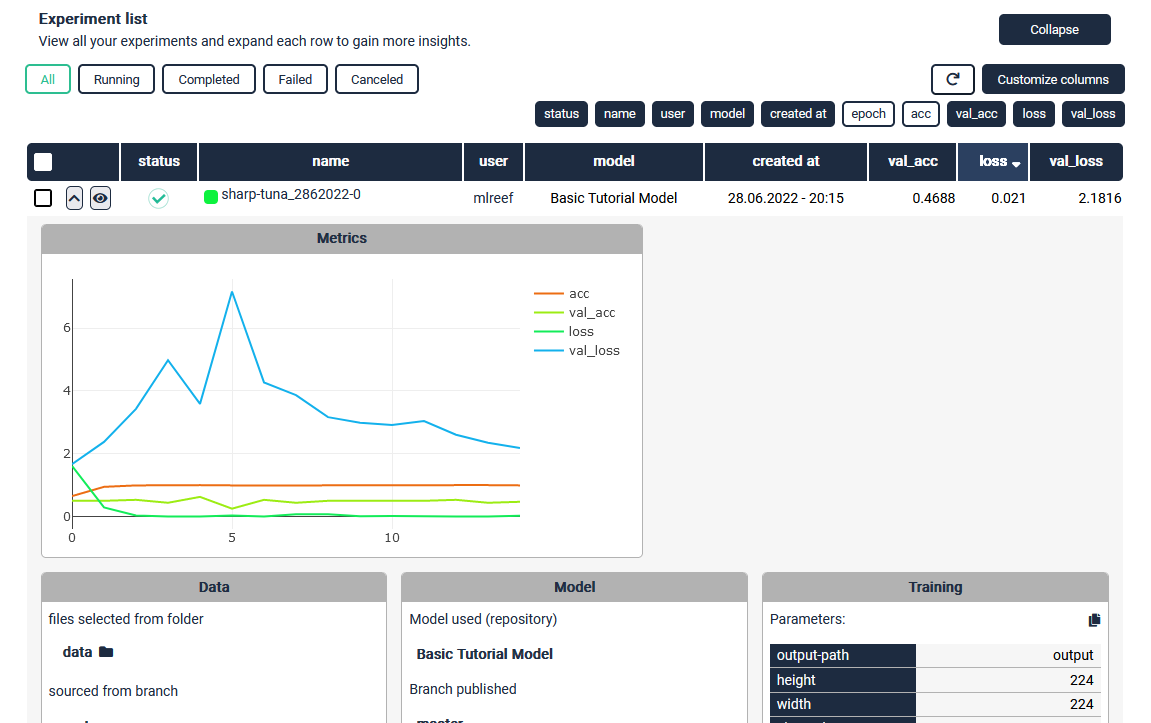
MLReef tracks and monitors your experiments. When you train a new model, you can see and review the metrics and performance via the experiment tracking section. In order to give MLReef the required information, you need to output within and "output" folder a experiment.json file.
For example, keras will create such a file with the below function:
class Metrics(keras.callbacks.Callback):
def on_train_begin(self, logs={}):
self.metrics = []
self.metrics2 = {}
with open('{}/experiment.json'.format(output_path), 'w') as file:
json.dump(self.metrics, file)
def on_batch_end(self, batch, logs={}):
try:
self.metrics2[batch] = {
'acc': float(logs.get('acc')),
'loss': float(logs.get('loss')),
}
with open('{}/experiment_batch.json'.format(output_path), 'w') as file:
json.dump(self.metrics2, file)
except Exception as identifier:
print("Error encountered: ", identifier)
return None
In MLReef, your experiment than will create after each epoch a new entry in your json file and MLReef will continuously read it. The result can look something like this: